Let's Get Numerical
A parabola is tangent to the line and to the line , and passes through the points (the green dots). Find the distance between the vertex of the parabola and its focus (the focal distance).
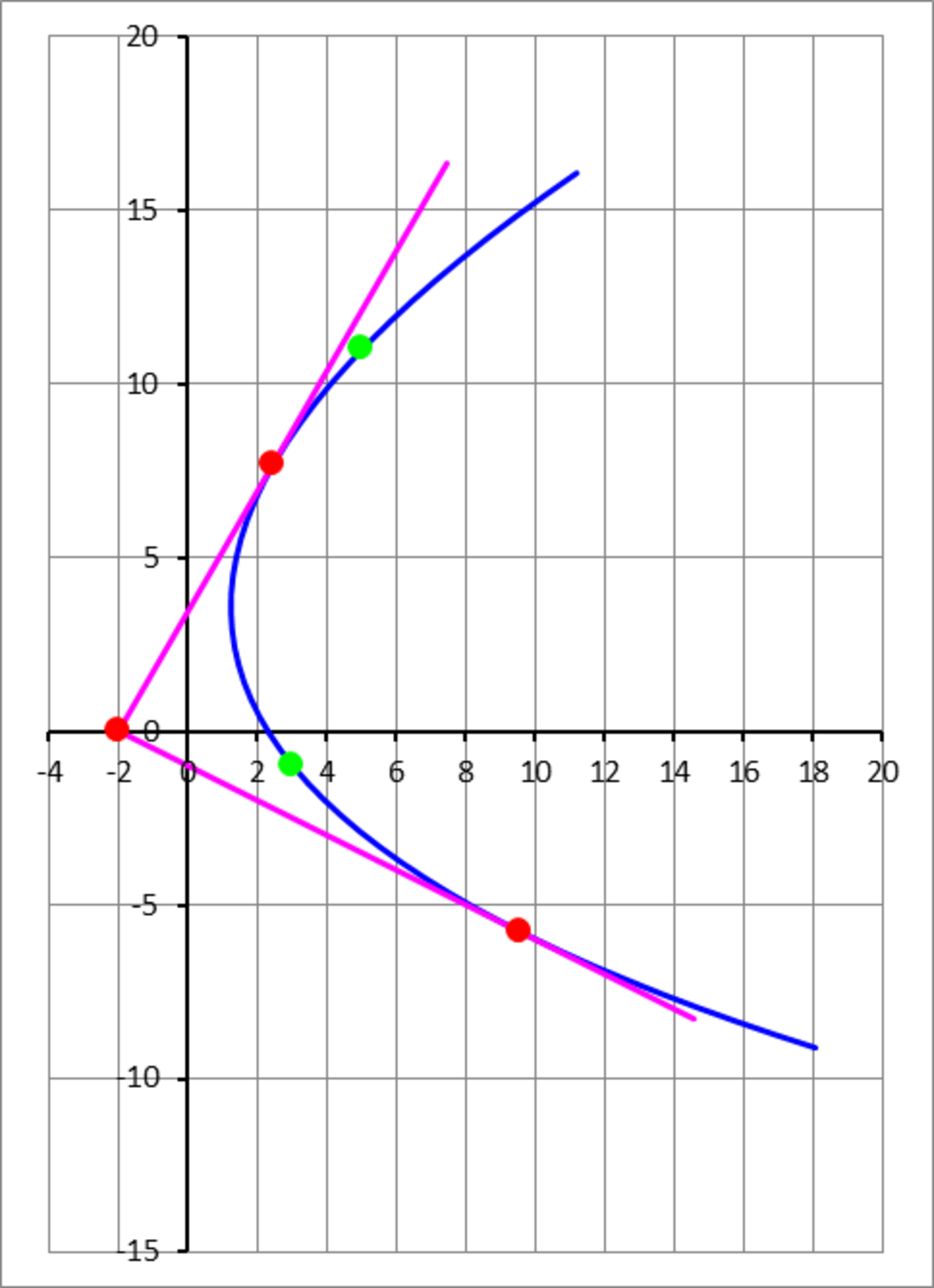
The answer is 3.1424.
This section requires Javascript.
You are seeing this because something didn't load right. We suggest you, (a) try
refreshing the page, (b) enabling javascript if it is disabled on your browser and,
finally, (c)
loading the
non-javascript version of this page
. We're sorry about the hassle.

Let P 0 be the vertex location, and let u 1 and u 2 be orthogonal unit vectors which effectively define a new coordinate system. Define points ( x 1 , y 1 ) = ( 3 , − 1 ) and ( x 2 , y 2 ) = ( 5 , 1 1 ) (the green points). The coordinates of a point on the parabola are:
P = P 0 + α u 1 + a α 2 u 2 x = x 0 + α u 1 x + a α 2 u 2 x y = y 0 + α u 1 y + a α 2 u 2 y u 1 = ( cos θ , sin θ ) u 2 = ( sin θ , − cos θ )
Each green point passed through has an associated α value. So the six unknowns are ( x 0 , y 0 , θ , a , α 1 , α 2 ) . We need to solve six equations for the six unknowns. The first four have to do with the coordinates of the two green points passed through.
x 1 = x 0 + α 1 u 1 x + a α 1 2 u 2 x y 1 = y 0 + α 1 u 1 y + a α 1 2 u 2 y x 2 = x 0 + α 2 u 1 x + a α 2 2 u 2 x y 2 = y 0 + α 2 u 1 y + a α 2 2 u 2 y
Consider the tangential intersection with the line y = 3 ( x + 2 ) . There should only be one corresponding α value. The quadratic equation for α should only have one solution. This gives us the fifth equation.
y = 3 ( x + 2 ) y 0 + α u 1 y + a α 2 u 2 y = 3 x 0 + 3 α u 1 x + 3 a α 2 u 2 x + 2 3 A = a u 2 y − 3 a u 2 x B = u 1 y − 3 u 1 x C = y 0 − 3 x 0 − 2 3 B 2 − 4 A C = 0
We can do the same thing for the tangential intersection with the line y = − 2 1 ( x + 2 ) , yielding the sixth equation.
y = − 2 1 ( x + 2 ) y 0 + α u 1 y + a α 2 u 2 y = − 2 1 x 0 − 2 1 α u 1 x − 2 1 a α 2 u 2 x − 1 A ′ = a u 2 y + 2 1 a u 2 x B ′ = u 1 y + 2 1 u 1 x C ′ = y 0 + 2 1 x 0 + 1 B ′ 2 − 4 A ′ C ′ = 0
I used a genetic algorithm to solve (with mutation plus hybridization). The GA has better convergence than a pure hill-climbing algorithm (with mutation only). Its convergence is not as good as that of a multi-variate Newton-Raphson algorithm, but it is simpler to implement, given the complexity of the equations involved. The focal length is equal to 4 a 1 , and it is approximately 3 . 1 4 . Approximate values for the other parameters are also given below.
Here is the Python code for the GA. Disclaimer: I'm not a programmer, so it probably looks pretty amateurish.